Scikit Image Example¶
A Python application that demonstrates how to use Scikit Image to apply filters to images based on a Qt Widgets.
import sys
import numpy as np
from matplotlib.backends.backend_qt5agg import FigureCanvas
from matplotlib.colors import LinearSegmentedColormap
from matplotlib.figure import Figure
from PySide6.QtCore import Qt, Slot
from PySide6.QtGui import QAction, QKeySequence
from PySide6.QtWidgets import (QApplication, QHBoxLayout, QLabel,
QMainWindow, QPushButton, QSizePolicy,
QVBoxLayout, QWidget)
from skimage import data
from skimage.color import rgb2hed
from skimage.exposure import rescale_intensity
class ApplicationWindow(QMainWindow):
"""
Example based on the example by 'scikit-image' gallery:
"Immunohistochemical staining colors separation"
https://scikit-image.org/docs/stable/auto_examples/color_exposure/plot_ihc_color_separation.html
"""
def __init__(self, parent=None):
QMainWindow.__init__(self, parent)
self._main = QWidget()
self.setCentralWidget(self._main)
# Main menu bar
self.menu = self.menuBar()
self.menu_file = self.menu.addMenu("File")
exit = QAction("Exit", self, triggered=qApp.quit)
self.menu_file.addAction(exit)
self.menu_about = self.menu.addMenu("&About")
about = QAction("About Qt", self, shortcut=QKeySequence(QKeySequence.HelpContents),
triggered=qApp.aboutQt)
self.menu_about.addAction(about)
# Create an artificial color close to the original one
self.ihc_rgb = data.immunohistochemistry()
self.ihc_hed = rgb2hed(self.ihc_rgb)
main_layout = QVBoxLayout(self._main)
plot_layout = QHBoxLayout()
button_layout = QHBoxLayout()
label_layout = QHBoxLayout()
self.canvas1 = FigureCanvas(Figure(figsize=(5, 5)))
self.canvas2 = FigureCanvas(Figure(figsize=(5, 5)))
self._ax1 = self.canvas1.figure.subplots()
self._ax2 = self.canvas2.figure.subplots()
self._ax1.imshow(self.ihc_rgb)
plot_layout.addWidget(self.canvas1)
plot_layout.addWidget(self.canvas2)
self.button1 = QPushButton("Hematoxylin")
self.button2 = QPushButton("Eosin")
self.button3 = QPushButton("DAB")
self.button4 = QPushButton("Fluorescence")
self.button1.setSizePolicy(QSizePolicy.Preferred, QSizePolicy.Expanding)
self.button2.setSizePolicy(QSizePolicy.Preferred, QSizePolicy.Expanding)
self.button3.setSizePolicy(QSizePolicy.Preferred, QSizePolicy.Expanding)
self.button4.setSizePolicy(QSizePolicy.Preferred, QSizePolicy.Expanding)
self.button1.clicked.connect(self.plot_hematoxylin)
self.button2.clicked.connect(self.plot_eosin)
self.button3.clicked.connect(self.plot_dab)
self.button4.clicked.connect(self.plot_final)
self.label1 = QLabel("Original", alignment=Qt.AlignCenter)
self.label2 = QLabel("", alignment=Qt.AlignCenter)
font = self.label1.font()
font.setPointSize(16)
self.label1.setFont(font)
self.label2.setFont(font)
label_layout.addWidget(self.label1)
label_layout.addWidget(self.label2)
button_layout.addWidget(self.button1)
button_layout.addWidget(self.button2)
button_layout.addWidget(self.button3)
button_layout.addWidget(self.button4)
main_layout.addLayout(label_layout, 2)
main_layout.addLayout(plot_layout, 88)
main_layout.addLayout(button_layout, 10)
# Default image
self.plot_hematoxylin()
def set_buttons_state(self, states):
self.button1.setEnabled(states[0])
self.button2.setEnabled(states[1])
self.button3.setEnabled(states[2])
self.button4.setEnabled(states[3])
@Slot()
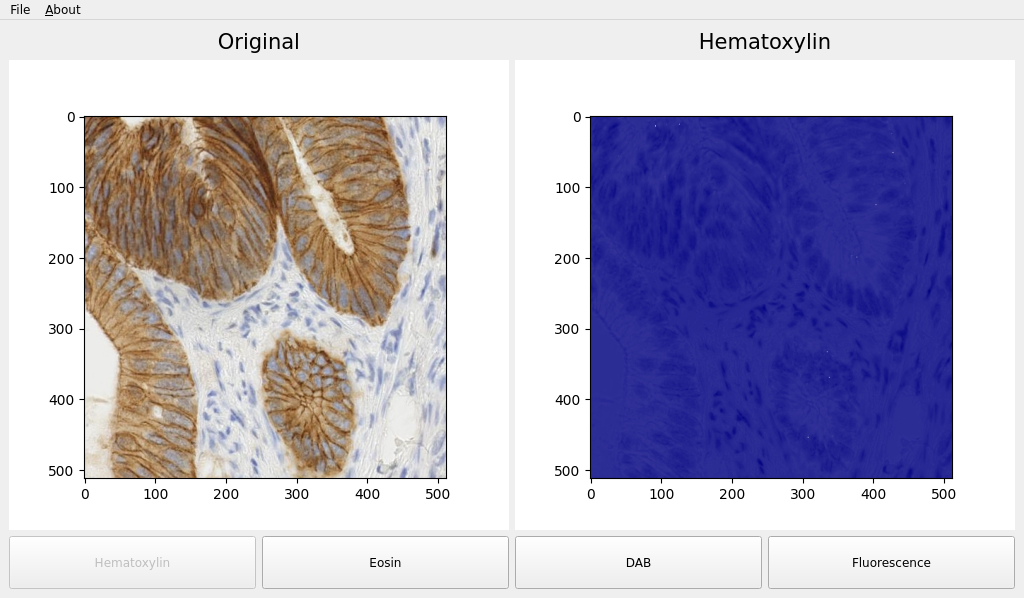
def plot_hematoxylin(self):
cmap_hema = LinearSegmentedColormap.from_list("mycmap", ["white", "navy"])
self._ax2.imshow(self.ihc_hed[:, :, 0], cmap=cmap_hema)
self.canvas2.draw()
self.label2.setText("Hematoxylin")
self.set_buttons_state((False, True, True, True))
@Slot()
def plot_eosin(self):
cmap_eosin = LinearSegmentedColormap.from_list("mycmap", ["darkviolet", "white"])
self._ax2.imshow(self.ihc_hed[:, :, 1], cmap=cmap_eosin)
self.canvas2.draw()
self.label2.setText("Eosin")
self.set_buttons_state((True, False, True, True))
@Slot()
def plot_dab(self):
cmap_dab = LinearSegmentedColormap.from_list("mycmap", ["white", "saddlebrown"])
self._ax2.imshow(self.ihc_hed[:, :, 2], cmap=cmap_dab)
self.canvas2.draw()
self.label2.setText("DAB")
self.set_buttons_state((True, True, False, True))
@Slot()
def plot_final(self):
h = rescale_intensity(self.ihc_hed[:, :, 0], out_range=(0, 1))
d = rescale_intensity(self.ihc_hed[:, :, 2], out_range=(0, 1))
zdh = np.dstack((np.zeros_like(h), d, h))
self._ax2.imshow(zdh)
self.canvas2.draw()
self.label2.setText("Stain separated image")
self.set_buttons_state((True, True, True, False))
if __name__ == "__main__":
app = QApplication(sys.argv)
w = ApplicationWindow()
w.show()
app.exec()


